This Mathematica notebook gives an overview of what can be done with my cellular automaton and correlation dynamics packages.
More info on the pages on correlation dynamics on my personal website.
Put the packages in Mathematica's preferred locations or place them together with the notebook. Modify the following so that Mathematica operates in the right directory
In[1]:=
![]()
In[2]:=
![]()
Out[2]=
![]()
In[3]:=
![]()
Out[3]=
![]()
We are going to define a model where every site is either empty, occupied by a healthy (sus) or an infected (inf) host:
In[4]:=
![]()
Out[4]=
![]()
The events that change the states of sites are
In[5]:=
![eventList := {event[ {oc[i_], emp}, {oc[i_], oc[sus]}] :> phi b[i], event[ {oc[i_], x_}, {emp, x_}] :> phi d[i], event[ {oc[sus], oc[inf]}, {oc[inf], oc[inf]}] :> phi beta, event[ {oc[i_], emp}, {emp, oc[i_]}] :> phi m[i]} ;](HTMLFiles/demo.nb_8.gif)
which represent reproduction, mortality, infection and movement.
In[6]:=
![parameterValues := {phi -> 1/6, (* note : phi equals one over the number of neighbours *) theta -> 2/5, b[sus] -> b0, b[inf] -> (1 - epsilon) b0, d[sus] -> d0, d[inf] -> d0 + alpha, m[sus] -> 2, m[inf] -> 4, b0 -> 4, d0 -> 1, beta -> 20, alpha -> 0.5, epsilon -> 0.75, init[emp] -> 1 - init[oc[sus]] - init[oc[inf]], init[oc[sus]] -> 0.6, init[oc[inf]] -> 0.001}](HTMLFiles/demo.nb_9.gif)
some definitions for displaying things:
In[7]:=
![]()
some simulation specifications
In[10]:=
![]()
In[11]:=
![]()
Out[11]=
![]()
In[12]:=
![]()
Out[12]=
![]()
In[13]:=
![]()
Out[13]//TableForm=
|
|
|
|
|
|
In[14]:=
![]()
In[15]:=
![]()
In[16]:=
![]()
In[17]:=
![pMF = Plot[ {p[oc[sus], mf[t]], p[oc[inf], mf[t]]}, {t, 0, tfin}, PlotStyle -> {{Dashing[{0.03, 0.01}], color[sus]}, {Dashing[{0.03, 0.01}], color[inf]}}, PlotRange -> {0, Automatic}]](HTMLFiles/demo.nb_23.gif)
![[Graphics:HTMLFiles/demo.nb_24.gif]](HTMLFiles/demo.nb_24.gif)
Out[17]=
![]()
In[18]:=
![]()
Global densities
In[21]:=
![pIPA = Plot[ {p[oc[sus], ipa[t]], p[oc[inf], ipa[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.02], light[color[sus]]}, {Thickness[0.02], light[color[inf]]}}, PlotRange -> {0, 0.6}]](HTMLFiles/demo.nb_27.gif)
![[Graphics:HTMLFiles/demo.nb_28.gif]](HTMLFiles/demo.nb_28.gif)
Out[21]=
![]()
Densities as `seen' by the parasite
In[22]:=
![qIPA = Plot[ {q[oc[sus], oc[inf], ipa[t]], q[oc[inf], oc[inf], ipa[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.02], light[color[sus]]}, {Thickness[0.02], light[color[inf]]}}, PlotRange -> {0, 0.6}]](HTMLFiles/demo.nb_30.gif)
![[Graphics:HTMLFiles/demo.nb_31.gif]](HTMLFiles/demo.nb_31.gif)
Out[22]=
![]()
In[23]:=
![]()
Out[23]=
![]()
In[24]:=
![]()
Out[24]=
![]()
In[25]:=
![]()
Out[25]=
![]()
In[26]:=
![]()
Out[26]=
![]()
In[27]:=
![]()
Out[27]=
![p[{oc[inf], oc[inf]}] (d[inf] + ihp m[inf] q[emp, {oc[inf], oc[inf]}]) + p[{oc[sus], oc[inf]}] (d[sus] + ihp m[sus] q[emp, {oc[sus], oc[inf]}]) + ihp m[inf] p[{emp, emp}] q[oc[inf], {emp, emp}] + beta ihp p[{emp, oc[sus]}] q[oc[inf], {oc[sus], emp}] + p[{emp, oc[inf]}] (-phi b[inf] - d[inf] - ihp m[inf] q[emp, {oc[inf], emp}] - ihp b[inf] q[oc[inf], {emp, oc[inf]}] - ihp m[inf] q[oc[inf], {emp, oc[inf]}] - ihp b[sus] q[oc[sus], {emp, oc[inf]}] - ihp m[sus] q[oc[sus], {emp, oc[inf]}])](HTMLFiles/demo.nb_42.gif)
These equations can be worked a bit more to look like proper correlation dynamics equations, but Mathematica remains rather limited in its capacity to represent symbolic output.
Define temporal...
In[28]:=
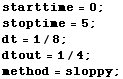
... and spatial aspects of the simulation
In[33]:=
![]()
where to store the results
In[36]:=
![]()
... and put together parameter files instructing the external module simpca
In[37]:=
![]()
![]()
![]()
In[38]:=
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
Out[38]=
![]()
(This takes a long time, but using Mathematica for this kind of simulation is like using a gun to kill a mosquito, whatever its original author might say)
Global and local densities:
In[39]:=
![pPCA1 = Plot[ {p[oc[sus], pca[t]], p[oc[inf], pca[t]]}, {t, 0, stoptime}, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_73.gif)
![[Graphics:HTMLFiles/demo.nb_74.gif]](HTMLFiles/demo.nb_74.gif)
Out[39]=
![]()
In[40]:=
![qPCA1 = Plot[ {q[oc[sus], oc[inf], pca[t]], q[oc[inf], oc[inf], pca[t]]}, {t, 0, stoptime}, PlotPoints -> 50, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_76.gif)
![[Graphics:HTMLFiles/demo.nb_77.gif]](HTMLFiles/demo.nb_77.gif)
Out[40]=
![]()
In[41]:=
![]()
Where to store the results
In[42]:=
![]()
... and put together parameter files instructing the external module simpca
In[43]:=
![]()
![]()
![]()
In[44]:=
![]()
![]()
![]()
![]()
![]()
In[45]:=
![]()
The following two commands read from file the simulation results
In[46]:=
![]()
In[47]:=
![]()
Global and local densities
In[48]:=
![pPCA2 = Plot[ {p[oc[sus], pca[t]], p[oc[inf], pca[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_92.gif)
![[Graphics:HTMLFiles/demo.nb_93.gif]](HTMLFiles/demo.nb_93.gif)
Out[48]=
![]()
In[49]:=
![qPCA2 = Plot[ {q[oc[sus], oc[inf], pca[t]], q[oc[inf], oc[inf], pca[t]]}, {t, 0, tfin}, PlotPoints -> 50, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_95.gif)
![[Graphics:HTMLFiles/demo.nb_96.gif]](HTMLFiles/demo.nb_96.gif)
Out[49]=
![]()
with mean field model
In[50]:=
![]()
![[Graphics:HTMLFiles/demo.nb_99.gif]](HTMLFiles/demo.nb_99.gif)
Out[50]=
![]()
with correlation dynamics model
In[51]:=
![]()
![[Graphics:HTMLFiles/demo.nb_102.gif]](HTMLFiles/demo.nb_102.gif)
Out[51]=
![]()
Now, compare predictions about the local environment of parasites
In[52]:=
![]()
![[Graphics:HTMLFiles/demo.nb_105.gif]](HTMLFiles/demo.nb_105.gif)
Out[52]=
![]()
In[53]:=
![]()
In[55]:=
![]()
Out[55]=
![]()
In[56]:=
![]()
![[Graphics:HTMLFiles/demo.nb_111.gif]](HTMLFiles/demo.nb_111.gif)
Out[56]=
![]()
In[57]:=
![]()
To show how vital rates can be density dependent, let us introduce an Allee effect
In[58]:=
![eventList := {event[ {oc[i_], emp}, {oc[i_], oc[sus]}] :> phi b[i] (phi 2 (1 - nb[emp, $focus]))^2, event[ {oc[i_], x_}, {emp, x_}] :> phi d[i], event[ {oc[sus], oc[inf]}, {oc[inf], oc[inf]}] :> phi beta, event[ {oc[i_], emp}, {emp, oc[i_]}] :> phi m[i]} ;](HTMLFiles/demo.nb_114.gif)
here it is assumed that birth rates increase with double the square of local density (phi is one over the number of neighbours)
In[59]:=
![]()
Global densities
In[62]:=
![pIPA3 = Plot[ {p[oc[sus], ipa[t]], p[oc[inf], ipa[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.02], light[color[sus]]}, {Thickness[0.02], light[color[inf]]}}, PlotRange -> {0, 0.6}]](HTMLFiles/demo.nb_116.gif)
![[Graphics:HTMLFiles/demo.nb_117.gif]](HTMLFiles/demo.nb_117.gif)
Out[62]=
![]()
Densities as `seen' by the parasite
In[63]:=
![qIPA3 = Plot[ {q[oc[sus], oc[inf], ipa[t]], q[oc[inf], oc[inf], ipa[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.02], light[color[sus]]}, {Thickness[0.02], light[color[inf]]}}, PlotRange -> {0, 0.6}]](HTMLFiles/demo.nb_119.gif)
![[Graphics:HTMLFiles/demo.nb_120.gif]](HTMLFiles/demo.nb_120.gif)
Out[63]=
![]()
In[64]:=
![]()
In[65]:=
![]()
![]()
![]()
In[66]:=
![]()
![]()
![]()
![]()
![]()
In[67]:=
![]()
In[68]:=
![]()
In[69]:=
![]()
Results
In[70]:=
![pPCA3 = Plot[ {p[oc[sus], pca[t]], p[oc[inf], pca[t]]}, {t, 0, tfin}, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_134.gif)
![[Graphics:HTMLFiles/demo.nb_135.gif]](HTMLFiles/demo.nb_135.gif)
Out[70]=
![]()
In[71]:=
![qPCA3 = Plot[ {q[oc[sus], oc[inf], pca[t]], q[oc[inf], oc[inf], pca[t]]}, {t, 0, tfin}, PlotPoints -> 50, PlotStyle -> {{Thickness[0.01], color[sus]}, {Thickness[0.01], color[inf]}}, PlotRange -> {0, 0.8}]](HTMLFiles/demo.nb_137.gif)
![[Graphics:HTMLFiles/demo.nb_138.gif]](HTMLFiles/demo.nb_138.gif)
Out[71]=
![]()
In[72]:=
![]()
![[Graphics:HTMLFiles/demo.nb_141.gif]](HTMLFiles/demo.nb_141.gif)
Out[72]=
![]()
In[73]:=
![]()
Out[73]=
![]()
In[74]:=
![]()
![[Graphics:HTMLFiles/demo.nb_146.gif]](HTMLFiles/demo.nb_146.gif)
Out[74]=
![]()
Converted by Mathematica (February 8, 2005)